| |
| Med Sci (Paris). 34: 52–58. doi: 10.1051/medsci/201834f110.Computational QSAR model combined molecular descriptors and fingerprints to predict HDAC1 inhibitors Jingsheng Shi,1 Guanglei Zhao,1 and Yibing Wei1* 1Division of Orthopaedic Surgery, Huashan Hospital, Fudan University, Shanghai, China |
The dynamic balance between acetylation and deacetylation of histones plays a crucial role in the epigenetic regulation of gene expression. It is equilibrated by two families of enzymes: histone acetyltransferases and histone deacetylases (HDACs). HDACs repress transcription by regulating the conformation of the higher-order chromatin structure. HDAC inhibitors have recently become a class of chemical agents for potential treatment of the abnormal chromatin remodeling process involved in certain cancers. In this study, we constructed a large dataset to predict the activity value of HDAC1 inhibitors. Each compound was represented with seven fingerprints, and computational models were subsequently developed to predict HDAC1 inhibitors via five machine learning methods. These methods include naïve Bayes, κ-nearest neighbor, C4.5 decision tree, random forest, and support vector machine (SVM) algorithms. The best predicting model was CDK fingerprint with SVM, which exhibited an accuracy of 0.89. This model also performed best in five-fold cross-validation. Some representative substructure alerts responsible for HDAC1 inhibitors were identified by using MoSS in KNIME, which could facilitate the identification of HDAC1 inhibitors. Keywords: QSAR, fingerprints, HDAC1 inhibitors |
Generally, reversible acetylation of lysine residues in the N-terminal tails of histone proteins significantly influences gene expression in all aspects of biology, such as cell proliferation, chromosome remodeling, and gene transcription [1]. Histone hyperacetylation facilitates gene expression, whereas histone deacetylation represses transcription. The dynamic balance of reversible acetylation of histones is guided by competitive activities of two corresponding enzymes, namely, histone acetyltransferases and histone deacetylases (HDACs). The family of human HDACs is categorized into four classes according to structure and function: class I (HDAC1, 2, 3, and 8), class II (HDAC4, 5, 6, 7, 9, and 10), class III (Sirt1, 2, 3, 4, 5, 6, and 7), which consists of NAD+-dependent proteins for deacetylation reaction, and class IV (HDAC11), which are zinc-dependent amido-hydrolases [2, 3]. Recent studies have revealed the potential of HDACs as novel therapeutic targets in reversing aberrant gene expression associated with certain cancers [4]. Deregulation of HDAC recruitment to target gene promoters can result in tumorigenesis, and overexpression of HDACs can mediate tumor cell proliferation [5]. HDAC inhibitors have also effectively repressed tumor growth in animal models of cancer [2, 6-9]. These inhibitors can arrest the cell cycle in the G1/G2 phase, thereby controlling tumor cell growth. These small molecule inhibitors also show a greater sensitivity to transformed cells compared with normal cells, thereby allowing high tumor-cell selective killing [10, 11]. In addition, cells treated with HDAC inhibitors can initiate extrinsic (death receptor) and intrinsic (mitochondrial) pathways to induce apoptosis [12]. Thus, HDAC inhibitors can provide a promising new insight in targeted anti-cancer therapy. To predict and identify possible HDAC inhibitors, we constructed a quantitative structure–activity relationship (QSAR) modeling of HDAC1 inhibitors, which is a routine method of identifying novel compounds and structures. On the basis of chemical structures and properties, QSAR methods can predict the biological activity and classification of compounds, which can provide a time-efficient method of performing animal verification experiments. Numerous QSAR methods, such as 3D modeling methods, have been developed to study HDAC inhibitory activity [13]. Zhao et al. used a GA-kNN method to predict the activity value of HDAC1 and HDAC6 inhibitors [14]. QSAR methods combined with other methods such as similarity searching [15], pharmacophore matching [16], and virtual screening [17], can also be used to predict and identify novel HDAC inhibitors. However, the models and the underlying mechanism cannot be easily explained by merely using individual or simple chemical descriptors. New molecular features or mixing multiple features have recently been integrated in building models. In this study, we collected high-quality diverse data from the literature and databases. Binary classification prediction models were subsequently developed using seven fingerprints combined with five machine learning methods. Five-fold cross-validation and external set validation were employed to determine the predictive ability of the models. Substructure alerts [14] of HDAC1 inhibitors were analyzed using the MoSS module in KNIME, thereby obtaining important patterns. |
Data collection and preparation A total of 2,344 human HDAC1 inhibitors in the training set were extracted from the Binding DB database [ 18]. An external validation set that contains 413 compounds was downloaded from the ChEMBL database [ 19]. HDAC1 inhibition was expressed as IC 50, the half-maximal (50%) inhibitory concentration of a substance. Compounds were classified as HDAC1 inhibitors if the IC 50 value obtained was lower than 10,000 nM; compounds were considered non-inhibitors if the IC 50 value was greater than 10,000 nM. The first dataset was obtained from the two different databases in the SMILES format, with duplicates excluded. Inorganic and metal ion-contained compounds were omitted. Salt chemicals were transformed to corresponding acids or bases. Detailed statistical descriptions of the entire HDAC1 inhibitors datasets are listed in Table 1. HDAC1 inhibitors were represented as 1 and HDAC1 non-inhibitors as 0 when building binary classification models.
Table 1
|
|
Inhibition |
Non-inhibition |
Total number |
| IC50 (nM) |
≤10000 |
>10000 |
|
|
| Compound |
Training set |
2060 |
284 |
2344 |
| External validation set |
334 |
79 |
413 |
|
| Total number |
|
2394 |
363 |
2757 |
Statistical data of the chemicals in the training and the external validation sets of HDAC1 inhibitors.
|
Chemical space and similarity analysis of datasets The chemical space distribution of the dataset was defined by molecular weight (MW) and Ghose-Crippen LogKow (ALogP). The structural diversity of the dataset was assessed by the average Tanimoto similarity indexes based on MDL public keys. The “Calculate Diversity Metrics” protocol in Discovery Studio was used to calculate the average molecular similarity of the datasets. Calculation of molecular fingerprints PaDEL-Descriptor [ 19] was used to calculate seven molecular fingerprints for each compound, including CDK fingerprint (FP, 1024 bits), CDK Extended fingerprint (Ext, 1024 bits), Estate fingerprint (Est, 79 bits), MACCS keys (Mac, 166 bits), PubChem fingerprint (Pub, 881 bits), substructure fingerprint (FP4, 307 bits), and Klekota-Roth fingerprint (KR, 4860 bits). These fingerprints are described in the original studies [ 20, 21]. Model building methods Five machine learning methods were used to build the models, namely, naïve Bayes (NB), k-nearest neighbor ( κNN), C4.5 decision tree (CT), random forest (RF), and support vector machine (SVM). The first four methods were performed in the Orange Canvas 2.0 (available for free at http://www.ailab.si/orange/). The SVM algorithm was performed in LIBSVM 3.16 [ 22] (available for free at http://www.csie.ntu.edu.tw/~cjlin/libsvm/). Naïve Bayes (NB) NB is a simple classification method based on Bayes’ rule for conditional probability [ 23]. For NB classifiers, the method generates the prior probabilities that are directly provided on the basis of the core function of Eq. (1) [ 24]. Default settings in Orange Canvas were used.
κ-Nearest Neighbor (κNN) κNN predicts a classification for test cases on the basis of the majority voting of its κ-nearest neighbors in the feature space [ 25]. Nearness is measured by the Euclidian distance metrics. In this study, the parameter of k was set to five. C4.5 decision tree (CT) Developed by Quinlan, C4.5 is an algorithm used to generate a decision tree [ 26]. At each node of the tree, C4.5 chooses the data attribute that most effectively separates its sample set into subsets enriched in one class or the other. The attribute with the highest normalized information gain is chosen to make the decision. The algorithm then recurses on the smaller sublists. In this study, all parameters used the default values in Orange Canvas. Random forest (RF) RF is an ensemble learning method developed by Breiman for classification and regression [ 27]. In this approach, each tree in the ensemble is formed by first selection at random and a small group of input coordinates (features or variables hereafter) to split on at each node. The best split is then calculated based on these features in the training set. The tree is expanded to its maximum without pruning. Support vector machine (SVM) SVM, which was originally developed by Vapnik [ 28] for pattern recognition, is a classifier that searches for a decision boundary - a hyperplane - that discriminates between two classes [ 29]. This approach presents a limitation: in many cases, classes cannot be separated by a hyperplane and a nonlinear decision surface is required. This weakness can be addressed with the use of SVM by mapping the data from the original input space into a feature space in which a linear separator can be found. This mapping was obtained through the Gaussian radial basis function kernel. The penalty parameter C and different kernel parameters g were tuned based on the training set by using a grid search strategy and five-fold cross-validation. This approach was applied to determine the SVM model with the optimal performance. Analysis of substructure alerts This study employed the MoSS node in KNIME [ 30] to search for frequently occurring substructure fragments in the datasets. Christian Borgelt’s MoSS implementation was employed. Four parameters of MoSS significantly influenced the results of this node. The reasonable values of the parameters were determined by trial and error. The “minimum focus support in %” value was set to eight, and the “minimum fragment size” value was set to five. For the other parameters, default values were used. Assessment of the model performance Five-fold cross-validation and external set validation were used to test the performance of the models. All models were evaluated by the number of true positives ( TP), true negatives ( TN), false positives ( FP), and false negatives ( FN). Sensitivity ( SE), specificity ( SP), and classification accuracy ( CA) were also calculated. Sensitivity refers to the proportion of predicted inhibition chemicals among all HDAC1 inhibitors. Specificity is defined as the proportion of detected non-inhibition chemicals among all non-inhibitors of HDAC1. The overall classification accuracy is the proportion of correctly classified chemicals. The equations are expressed as follows: SE = TP/(TP + FN) (2) SP = TN/(TN + FP) (3) CA = (TP + TN)/(TP + TN + FP + FN)(4) The receiver operating characteristic curve was also determined to evaluate the quality of the binary classification model. If the plot has a surface area equal to one, then the classifier is perfect; if the area is equal to 0.5, then the classifier has no discriminative power at all [31, 32]. |
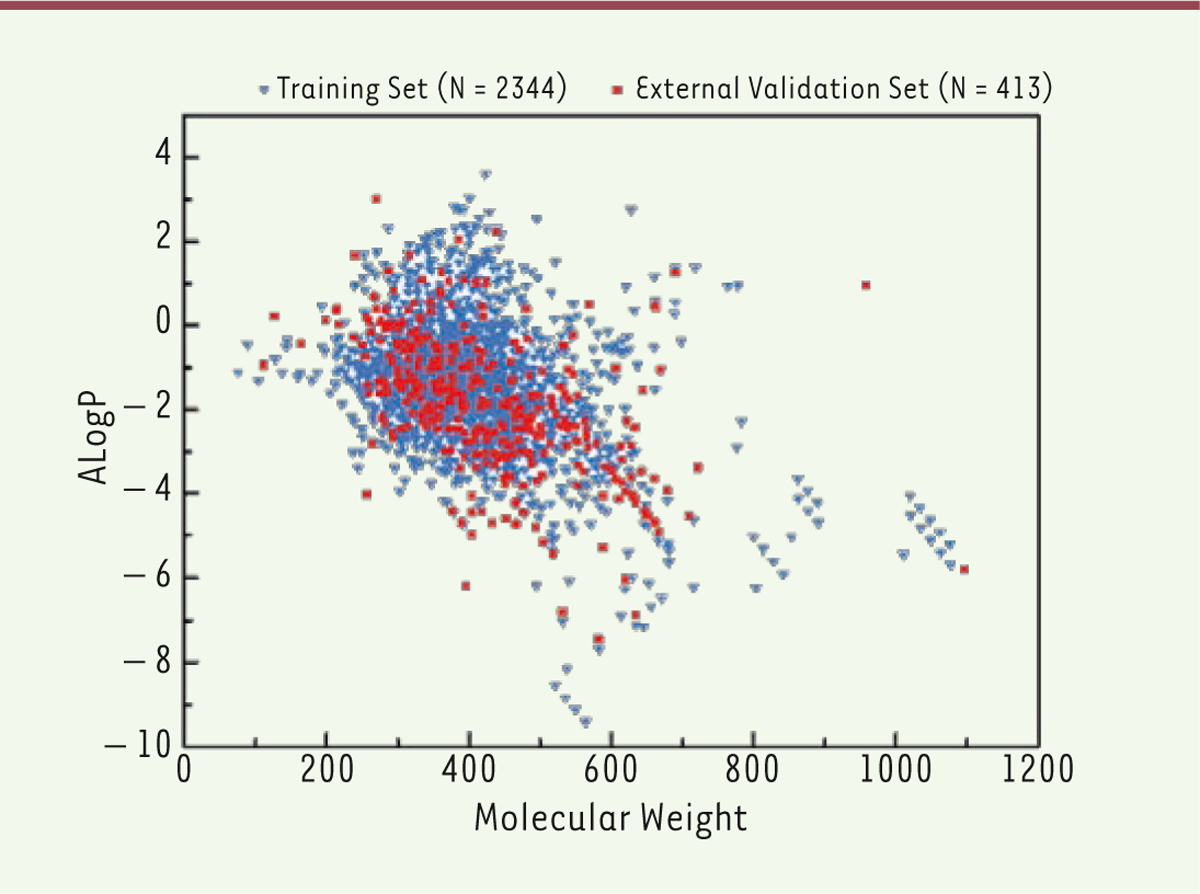
Chemical diversity analysis The number of the chemicals in the training set and in the external validation set of HDAC1 inhibitors were 2,344 and 413, respectively, as shown in Table 1. Chemical diversity is important to build a global and robust QSAR model. Therefore, we used the chemical space and Tanimoto similarity to investigate chemical diversity. The MW and ALogP of each class in the datasets were analyzed. The chemical space distribution plots of the datasets are depicted in Figure 1. The compounds of the training set of HDAC1 inhibitors are intensively distributed in the lower left region of the upper plot. The external validation set shares a similar chemical space with that of the training set. Several compounds recorded high molecular weights or high ALogP values, and these inhibitors can be hardly absorbed by humans. Therefore, they were deleted prior to the development of the models.
 | Figure 1. Chemical diversity analysis of training and external validation sets of HDAC1 inhibitors (N1 = 2344, N2 = 413). N represents the number of compounds in different datasets. Chemical space defined by MW and ALogP. |
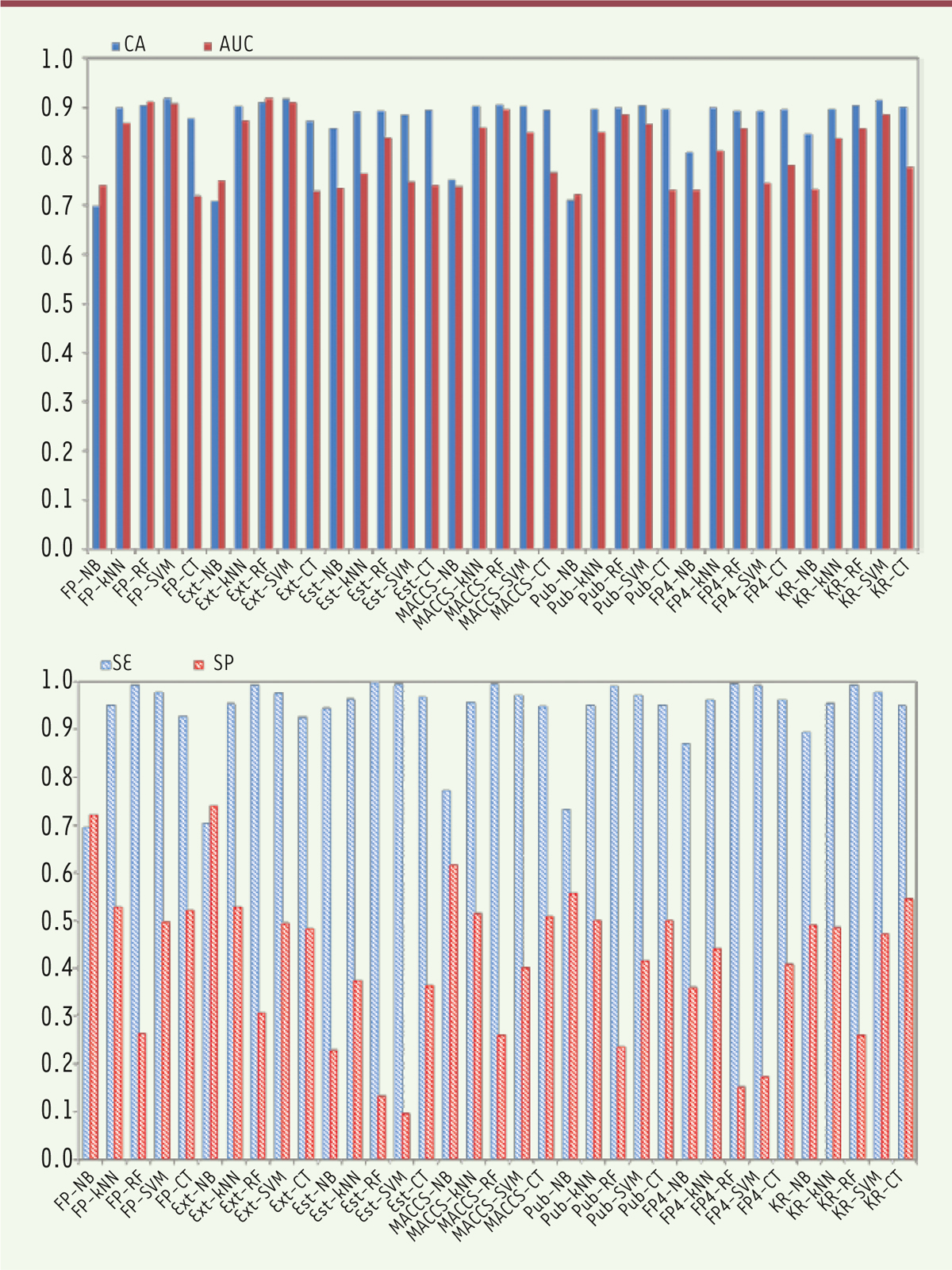
The average Tanimoto similarity indexes were calculated as 0.79 for the training set of the HDAC1 inhibitors and 0.73 for the external validation set. The average Tanimoto similarity index for the entire HDAC1 inhibitor dataset was determined to be 0.77, thereby suggesting chemical diversity. Performance of binary classification models Binary classification prediction models were developed using seven fingerprints combined with five machine learning methods, namely, NB, kNN, CT, RF, and SVM. The models were validated by five-fold cross-validation and external set validation. CA, SE, SP, and AUC values for the five-cross validation are summarized in Figure 2.
 | Figure 2. Performance of five-fold cross-validation for the training set of HDAC1 inhibitors. NB, naïve Bayes; k NN, k-nearest neighbor; CT, C4.5 decision tree; RF, random forest; SVM, support vector machine; FP, CDK fingerprint; Ext, CDK extended fingerprint; Est, estate fingerprint; MACCS, MACCS keys; Pub, PubChem fingerprint; FP4, substructure fingerprint; KR, Klekota-Roth fingerprint. The X-axis lists the name of the building models, and the Y-axis represents the values of CA, AUC, SE, SP. |
Five-fold cross-validation Five-fold cross-validation of the training set was performed to evaluate the robustness of the models. The optimal models were selected based on the CA and AUC values. The FP-SVM model ( CA = 0.91, SE = 0.97, SP = 0.50, AUC = 0.91) was chosen as the optimal model for HDAC1 inhibitors. Figure 2 (left histogram) reveals CA and AUC values of all prediction models exceeding 0.6. Figure 2 (right histogram) presents SE values that exceed those of SP values for most models, except for the FP-NB and Ext-NB models. Among the five machine learning methods using the same fingerprint, SVM and RF performed better than did the other three. Among the seven fingerprints using the same algorithm, FP and Ext obtained the most optimal results, whereas the predictive accuracies of the models using Est obtained the least optimal outcome. External set validation External validation sets were used to evaluate the predictive ability of the six best models from five-fold cross-validation. The results of the external set validation are listed in Table 2.
Table 2
| Rankinga |
HDAC1 |
CA |
SE |
SP |
AUC |
| 1 |
FP-SVM |
0.887 |
0.952 |
0.757 |
0.893 |
|
| 2 |
Ext-SVM |
0.886 |
0.955 |
0.600 |
0.879 |
|
| 3 |
Ext-RF |
0.818 |
0.997 |
0.075 |
0.879 |
|
| 4 |
FP-RF |
0.825 |
0.991 |
0.137 |
0.886 |
|
| 5 |
MACCS-RF |
0.823 |
0.976 |
0.185 |
0.836 |
|
| 6 |
KR-SVM |
0.862 |
0.912 |
0.650 |
0.866 |
Results of the external validation set with the top six models of the five-fold cross-validation. HDAC1, histone deacetylase 1 inhibitors; CA, classification accuracy; SE, sensitivity; SP, specificity; AUC, area under the receiver operating characteristic curve; FP, CDK Fingerprint; Ext, CDK extended fingerprint; MACCS, MACCS keys; KR, Klekota-Roth fingerprint; SVM, support vector machine; RF, random forest. |
As shown in Table 2, the best model for the HDAC1 inhibitors is FP combined with SVM algorithm (CA = 0.88, SE = 0.95, SP = 0.75, AUC = 0.89). This model also performed best in the five-fold cross-validation. The other five models are Ext-SVM, Ext-RF, FP-RF, MACCS-RF, and KR-SVM. The rankings of these five models for the HDAC1 inhibitors differed from their rankings in the five-fold cross-validation. For example, the predictive ability of the HDAC1 inhibitors MACCS-SVM in the five-fold cross-validation was higher than that in the FP-RF model. However, the latter performed much better than the former model in the external set validation. Substructure alerts of HDAC1 inhibitors Substructure alerts were analyzed for the entire datasets (including the training set and the external validation set) by using MoSS in KNIME. According to the results, 30 fragments were obtained for HDAC1 inhibitors. In this study, only eight selected typical fragments, shown in Table 3, are discussed. These fragments more frequently occur in HDAC1 inhibitors than in non-inhibitors, thereby indicating that a chemical that contains these substructure fragments is more likely to inhibit HDAC1.
Table 3
|
|
Structure fragment |
Support in focus (abs)a
|
Support in complement (abs)b
|
Support in focus (rel)c
|
Support in complement (rel)d
|
| 1 |
|
492 |
12 |
0.205 |
0.032 |
|
| 2 |
|
408 |
17 |
0.17 |
0.046 |
|
| 3 |
|
332 |
17 |
0.138 |
0.046 |
|
| 4 |
|
284 |
14 |
0.118 |
0.038 |
|
| 5 |
|
285 |
9 |
0.119 |
0.024 |
|
| 6 |
|
287 |
18 |
0.119 |
0.049 |
|
| 7 |
|
377 |
7 |
0.157 |
0.019 |
|
| 8 |
|
248 |
13 |
0.103 |
0.035 |
Substructural fragments of HDAC1 inhibitors obtained in MoSS. |
|
Diversity of the dataset Dataset diversity is a recognized as a key factor in QSAR modeling. A number of QSAR methods were developed to study HDAC inhibitors because of their satisfactory inhibitory activity against HDACs. Studies previously focused on the modeling of a class of HDAC inhibitors, and inhibitors against HDAC1 were mainly local QSAR modeling studies based on the chemical category 2D-QSAR, 3D-QSAR or pharmacophore [10,11]. However, these models provided reliable predictions only within a limited chemical space. Thus, we collected diverse data from the literature and the database to build global models. We used the chemical space and Tanimoto similarity index to investigate chemical diversity. The training sets shared a similar chemical space with the external validation sets, as shown in Figure 1. Some compounds were deleted because they have high molecular weights or high ALogP values; these compounds are hardly absorbed by humans. Moreover, the average Tanimoto similarity index of the entire dataset was 0.77, thereby indicating that our dataset was diverse to a certain extent. We constructed a classification model for predicting HDAC inhibitors on the basis of a large heterogeneous dataset of 2,344 compounds with high prediction accuracy. These values suggest that our models showed good generalization ability. Performance of binary classification models Five machine learning methods combined with seven fingerprints were used to build binary classification models for predicting HDAC1 inhibitors. Seven fingerprints were used to build models, unlike the traditional QSAR and pharmacophore models, which are built on molecular descriptors. Most of these traditional models were built by statistic methods with limited compounds and molecular descriptors. Here, we used fingerprints as attributes for QSAR modeling based on high-quality diverse data from the literature and databases. Fingerprints were good methods for chemical toxicity prediction and chemical metabolic property prediction which always made a direct connection between the chemical structure and toxicity endpoint of global compounds [ 33, 34]. In our study, we distinguish novel HDAC1 inhibitors and non-inhibitors by five machine learning methods combined with seven fingerprints. Figure 2 shows that SE values exceed SP values for most models, except for the FP-NB and Ext-NB models. In general, SVM and RF performed better than the three other algorithms when the same fingerprint was used. This result indicates that SVM and RF may be efficient methods to predict HDAC1 inhibitors. In addition, our method directly linked the chemical structure and the inhibition endpoint of compounds. The length of Est fingerprint is 79 bits, which may be inadequate to characterize chemical diversity. Given the same algorithm, FP and Ext obtained the best results among the seven fingerprints, whereas the models using Est obtained lower predictive accuracies. The selection of suitable fingerprints is important to characterize an entire dataset. Therefore, the best model for the HDAC1 inhibitors was FP combined with SVM algorithm, as shown in Table 2. In conclusion, we can use this model for further prediction of HADC1 inhibitor which was also performed best in the five-fold cross validation. Visualization analysis of substructure alerts In our research, we used the MoSS node in KNIME to analyze the substructure features of HDAC inhibitors. The substructure fragments were identified in the datasets which contain the HDAC1 inhibitors and non-inhibitors. According to the MoSS results, we obtained eight fragments that showed high frequency in HDAC1 inhibitors. These fragments exhibited two or three times greater frequency in inhibitors than in non-inhibitors. For example, the structure of vorinostat, an known HDAC inhibitor approved by the FDA for the treatment of cutaneous T-cell lymphoma, contains a chemical substructure of hydroxamates (fragment 2 in Table 3) [ 17]. Notably, several inhibitor compounds increase efficacy or reduce side effects because they contain certain chemical substructures (hydroxamates or benzamides) [ 35]. The results of MoSS in KNIME reveal that both hydroxamates and benzamides were specific fragments of HDAC1 inhibitors, which is consistent with previous results [ 36, 37]. Thus, our findings indicate that if a chemical contains these substructure fragments, then it exhibits greater potential as an HDAC1 inhibitor and can be more beneficial in the identification of the novel HDAC1 inhibitors. |
Seven fingerprints combined with five machine learning methods were used for modeling and predicting HDAC1 inhibitors. The best model for HDAC1 inhibitors was determined based on the CA and AUC values. The FP-SVM model obtained the highest prediction performance. Our study demonstrates that fingerprints can be used as attributes for QSAR modeling. Nevertheless, different fingerprints and algorithms fit different datasets, and the quality of the dataset directly affects the performance of the model. A substructure frequency was analyzed to identify substructure alerts that could be used to distinguish HDAC1 inhibitors from non-inhibitors. Finally, eight substructure fragments were identified, which pharmaceutical chemists can use to design and discover other potential HDAC1 inhibitors. Overall, this study demonstrates that machine learning methods and fingerprints can be used for in silico prediction of HDAC inhibitors, and that substructure fragment analysis can characterize the molecular features of HDAC inhibitors. Binary classification models can be applied in the screening of HDAC1 inhibitors in a time- and cost-efficient manner. |
Jingsheng Shi and Guanglei Zhao contribute equally to this work. The authors have no potential conflict of interests. |
Project supported by the National Natural Science Foundation of China (Grant No.81401829 )
|
1. Hu E, et al. Identification of novel isoform-selective inhibitors within class I histone deacetylases . J Pharmacol Exp Ther. 2003; ; 307 : :720.–728. 2. Bertrand P.. Inside HDAC with HDAC inhibitors . Eur J Med Chem. 2010; ; 45 : :2095.–2116. 3. Auzzas L, et al. Non-natural macrocyclic inhibitors of histone deacetylases: design, synthesis, and activity . J Med Chem. 2013; ; 53 : :8387.–8399. 4. Bolden JE, et al. Anticancer activities of histone deacetylase inhibitors . Nat Rev Drug Discov. 2006; ; 5 : :769.–784. 5. Kouzarides T.. Histone acetylases and deacetylases in cell proliferation . Curr Opin Genet Dev. 1999; ; 9 : :40.–42. 6. Grant S, Easley C, Kirkpatrick P. Vorinostat . Nat Rev Drug Disc Discov. 2007; ; 6 : :21.–22. 7. Huang L.. Targeting histone deacetylases for the treatment of cancer and inflammatory diseases . J Cell Physiol. 2006; ; 209 : :611.–616. 8. Liu T, Kuljaca S, Tee A, Marshall GM. Histone deacetylase inhibitors: multifunctional anticancer agents . Cancer Treat Rev. 2006; ; 32 : :157.–165. 9. Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic and more treatments for cancer . Nat Rev Cancer. 2006; ; 6 : :38.–54. 10. Qui L, et al. Anti-tumour activity in vitro and in vivo of selective differentiating agents containing hydroxamate . Br J Cancer. 1999; ; 80 : :1252.–1258. 11. Parsons PG, et al. Tumour selectivity and transcriptional activation by azelaic bishydroxamic acid in human melanocytic cells . Biochem Pharmacol. 1997; ; 53 : :1719.–1724. 12. Ma X, et al. Histone deacetylase inhibitors current status and overview of recent clinical trials . Drugs. 2009; ; 69 : :1911.–1934. 13. Tang H, et al. Combinatiorial QSAR modeling of specificity and subtype selectivity of ligands binding to serotonin receptors 5HT1E and 5HT1F . J Chem Inf Model. 2009; ; 49 : :461.. 14. Zhao LL, Xiang YH, Song JL, Zhang ZY. A novel two-step QSAR modeling work flow to predict selectivity and activity of HDAC inhibitors . Bioorg Med Chem Lett. 2003; ; 23 : :929.–933. 15. Juvale DC, et al. 3D-QSAR of histone deacetylase inhibitors: hydroxamate analogues . Org Biomol Chem. 2006; ; 4 : :2858.–2868. 16. Xiang YH, Hou ZY, Zhang ZY. Pharmacophore and QSAR studies to design novel histone deacetylase 2 inhibitors . Chen Biol Drug Des. 2012; ; 79 : :760.–770. 17. Hou X, et al. Enhancing the sensitivity of pharmacophore-based virtual screening by incorporating customized ZBG features: a case study using histone deacetylase 8 . J Chem Inf Model. 2005. 20. Yap CW. PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints . J Comput Chem. 2011; ; 32 : :1466.–1474. 21. Klekota J, Roth FP. Chemical substructures that enrich for biological activity . Bioinformatics. 2008; ; 24 : :2518.–2525. 22. Chang CC, Lin CJ. LIBSVM: a library for support vector machines . ACM Trans Intell Syst Technol. 2011; ; 2 : :27.. 23. Plewczynski D, Spieser SA, Koch U. Assessing different classification methods for virtual screening . J Chem Inf Model. 2006; ; 46 : :1098.–1106. 24. Watson P.. Naive Bayes classification using 2D pharmacophore feature triplet vectors . J Chem Inf Model. 2008; ; 48 : :166.–178. 25. Kauffman GW, Jurs PC. QSAR and k-nearest neighbor classification analysis of selective cyclooxygenase-2 inhibitors using topologically-based numerical descriptors . J Chem Inf Comput Sci. 2011; ; 41 : :1553.–1560. 26. Quinlan JR. C4.5: programs for machine learning . Morgan Kaufmann Publishers; Inc: San Francisco, CA: , 1993. 27. Breiman L.. Random forests . Machine Learning. 2001; ; 45 : :5.–32. 28. Cortes C, Vapnik V. Support-vector networks . Mach Learn. 1995; ; 20 : :273.–297. 29. Burges CJ. A tutorial on support vector machines for pattern recognition . Data Min. Knowl Discov. 1998; ; 2 : :121.–167. 31. Li J, Gramatica P. Classification and virtual screening of androgen receptor antagonists . J Chem Inf Model. 201050, :861.–874. 32. Li J, Gramatica P. QSAR classification of estrogen receptor binders and prescreening of potential pleiotropic EDCs . SAR QSAR Environ Res. 2010; ; 21 : :657.–669. 33. Chen YJ, Cheng FX, Sun L, et al. Computational models to predict endocrine-disrupting chemical binding with androgen or oestrogen receptors . Ecotoxicology and Environmental Safety. 2014; ; 110 : :280.–287. 34. Cheng F, Ikenaga Y, Zhou Y, et al. In silico assessment of chemical biodegradability . J Chem Inf Model. 2012; ; 52 : :655.–669. 35. Wanger JM, Hackanson B, Lubbert M, Jung M. Histone deacetylase(HDAC) inhibitors in recent clinical trials for cancer therapy . Clin Epigenet. 2010; ; 1 : :117.–136. 36. Tang H, Wang X, Huang X, et al. Novel inhibitors of human histone deacetylase (HDAC) identified by QSAR modeling of known inhibitors, virtual screening, and experimental validation . J Chem Inf Model. 2009; ; 49 : :461.–476. 37. Robey R, Chakraborty A, Basseville A, et al. Histone deacetylase inhibitors: emerging mechanisms of resistance . Mol Pharm. 2011; ; 8 : :2021.–2031. |